B-Store Datastores¶
| Author: | Kyle M. Douglass |
|---|---|
| Contact: | kyle.m.douglass@gmail.com |
| organization: | École Polytechnique Fédérale de Lausanne (EPFL) |
| revision: | $Revision: 1 $ |
| date: | 2017-01-24 |
| abstract: | The logic behind B-Store datastores is presented in this document. The HDF file type is briefly explained, followed by the organization of data within the database. |
Table of Contents
Introduction to B-Store Datastores¶
A single high-throughput SMLM experiment can generate hundreds or even thousands of different files containing different types of data. Analyzing this data requires that the files are sorted and organized in a well-structured way that is understandable by both humans and machines. A B-Store datastore fulfills this role as a structured container for heterogeneous SMLM data.
In basic terms, a B-Store datastore is a collection of individual datasets. Each dataset possesses identifiers that uniquely identify it within the datastore. A dataset also provides a container for the actual experimental data that it is holding, such as localizations or widefield images.
Datasets¶
A Dataset is a single, generalized dataset that can be stored in a B-Store datastore. It is “general” in the sense that it can represent one of a few different types of data (e.g. localizations, metadata, or widefield images). A specific type of dataset is called a DatasetType. A DatasetType knows what readers it may use to read raw input files from the disk and how to get and put data of its own type from and into a datastore. Unlike the Datastore, which sorts and organizes Datasets, the DatasetType encapsulates all the knowledge about data input and output.
There are currently five dataset types (examples of their raw input are in paranetheses):
- Localizations (tabulated localization data in raw text, csv format)
- LocMetadata (information about how the localizations were generated in JSON format)
- WidefieldImage (gray scale images; contains OME-XML metadata and Micro-Manager metadata)
- FiducialTracks (localizations belonging to individual fiducials in csv format)
- AverageFiducial (the average drift trajectory from many fiducials)
If you require a raw input type or a general DatasetType that is not listed here, B-Store can be easily extended to support it. Please let us know on the forum.
Dataset IDs¶
A dataset is uniquely defined by the following fields (the first four–prefix, acqID, datasetType, attributeOf–are required).
- prefix
- A descriptive name given to the dataset.
- acqID
- An integer that specifies the acquisition number of the dataset.
- datasetType
- A string. Must be one of the types listed above. The a type must be in the list __Registered_DatasetTypes__ in config.py to be used.
- attributeOf
- A string. Must be one of the dataset types listed above. This is the name of a type of dataset that this one describes.
- channelID
- (optional) A string that specifies the fluorescence channel that the dataset was acquired in.
- dateID
- (optional) A string in the format YYYY-MM-DD. This is for identifying the same field of view taken on different days.
- posID
- (optional) A one or two-element tuple of integers specifying the position of the field of view of the dataset.
- sliceID
- (optional) An integer identifying the the axial slice of the dataset.
- replicateID
- (optional) An integer identifying a replicate or biological repeat. This is used when a dataset has the same IDs as another but comes from an independent sample.
Hierarchy of Dataset IDs¶
All datasets with the same prefix are organized into the same acquisition group. Within an acquisition group, datasets are specified according to their acqID.
For example, let’s say we take three widefield images of Cos7 cells from the same coverslip during the same experiment. In the datastore, each image will have the same prefix, such as ‘Cos7’. The individual images however will have three different acqID’s. (Most likely they will be 1, 2, and 3, but they need not start at 1 or be sequential.)
If two datasets have the same prefix and acqID but different datasetType’s, then they will be understood to have come from the same field of view. This allows widefield images to be grouped with their corresponding localizations within the database. As an example, we might have two datasets in our datastore where both have ‘HeLa’ as a prefix and 1 as the acqID, but one has ‘Localizations’ as its datasetType and the other ‘WidefieldImage’.
Finally, the optional identifiers can further divide datasets that have the same prefix, acqID, and datasetTypes.
A diagram that explains this hierarchy is seen below. On top, you have your raw data files as inputs to a parser, which both assigns dataset IDs based on the files’ filename. A Reader converts the data into a format suitable for insertion into the database. A single acquisition group is identified by a prefix. Within this group, each dataset has a unique acqID and datasetType to set it apart from other datasets within the same group. Finally, the other optional IDs give you more control over how the data is organized within the group.
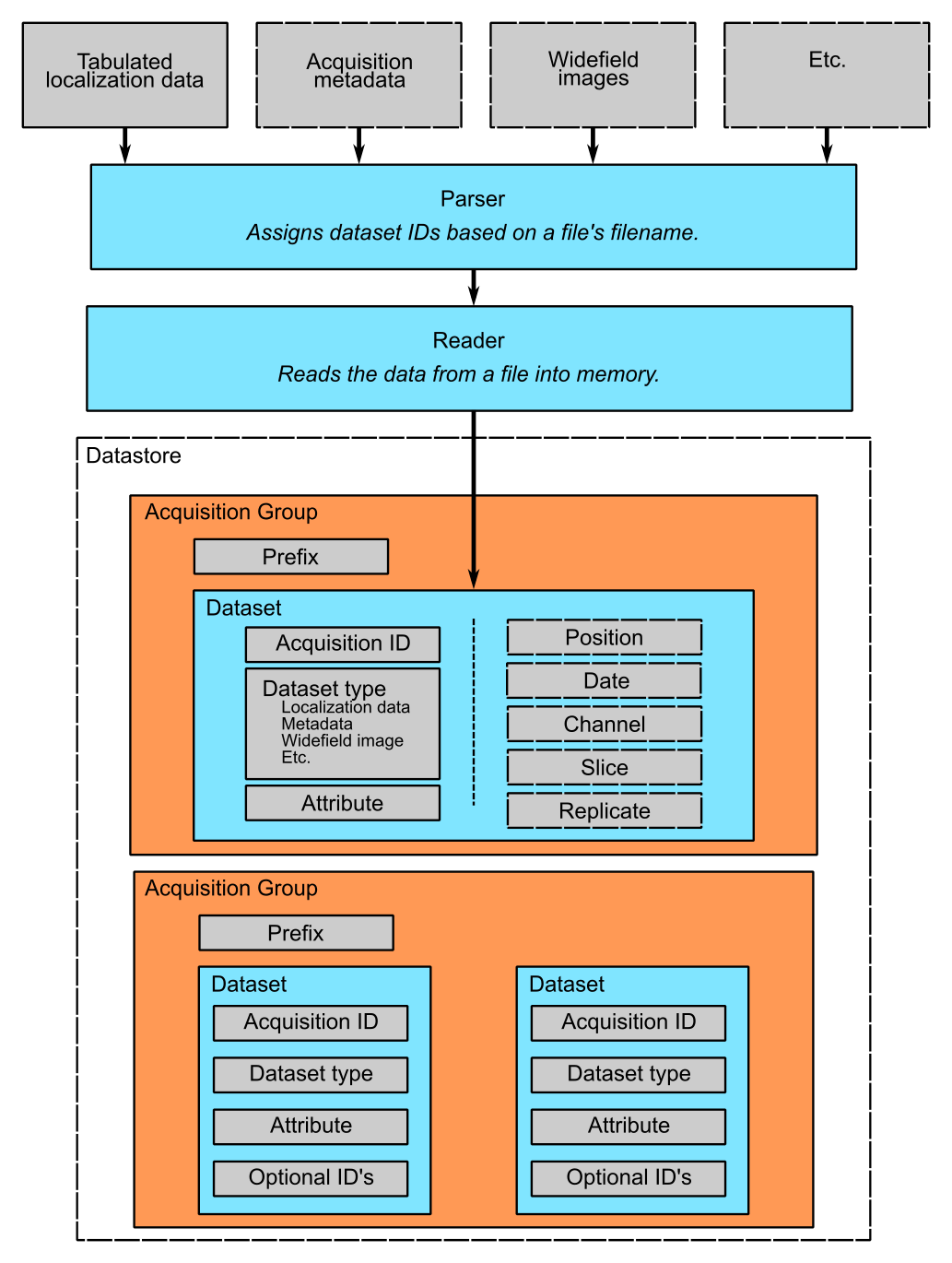
The Role of Parsers in Datastores¶
As mentioned above, a B-Store parser is an object that assigns dataset IDs to a dataset based on the filename of the file containing the data.
Since different labs often have very different ways to generate their data, parsers were designed to be very flexible objects. The only requirement of a parser is that it implements the functions described by the Parser metaclass; these functions specify the kinds of outputs a Parser must provide. The types of inputs, however, are not specified. This means that you can write a parser to convert any type of data that you would like into a dataset (as long as it fits within one of the datasetTypes). Furthermore, exactly how dataset IDs are assigned remains up to you. If you want your parser to label every single dataset with a prefix of ‘Bob’ then you can do that, though obviously the utility of such a feature will be in question.
This flexibility comes at a cost, however. If the built-in parsers do not work for your data, then it will be necessary to write your own. An example of how to do this is provided as a Jupyter notebook example.
The Role of Readers in Datastores¶
Readers do the actual work of reading the data inside a file into memory. When building a Datastore, a different reader may be specified for each dataset to allow B-Store to read data from a large range of file formats. Generic readers like CSVReader and JSONReader are provided for reading from generic file formats.
HDF Datastores¶
The HDFDatastore class allows for the creation of a datastore inside a HDF container. HDF is a high-performance file type used in scientific and numerical computing. It is considered a standard file type in scientific circles and is widely supported by many programming environments. One advantage of HDF containers is that you are not required to use B-Store code to access the data in a B-Store datastore. Any software that can read or modify HDF files will do.
HDFDatastore objects support many features of Python sets, like list comprehensions, filtering, and iteration.
HDFView¶
HDFView is a useful utility for viewing the contents of a HDF container. It is freely available and recommended for trouble shooting.
We will use screenshots taken from HDFView to explain how data is sorted inside a B-Store datastore.
Organization within an HDF datastore¶
The figure below is a screenshot from HDFView of the B-Store test database located in test_experiment/test_experiment_db.h5 in the B-Store test files repository. On left side of the window, you can see a hierarchy of the groups stored inside this database. There are two acqusition groups with prefixes HeLaL_Control and HeLaS_Control. Inside the HeLaL_Control group, you can see that there is one single acquisition (labeled with an acqID of 1).
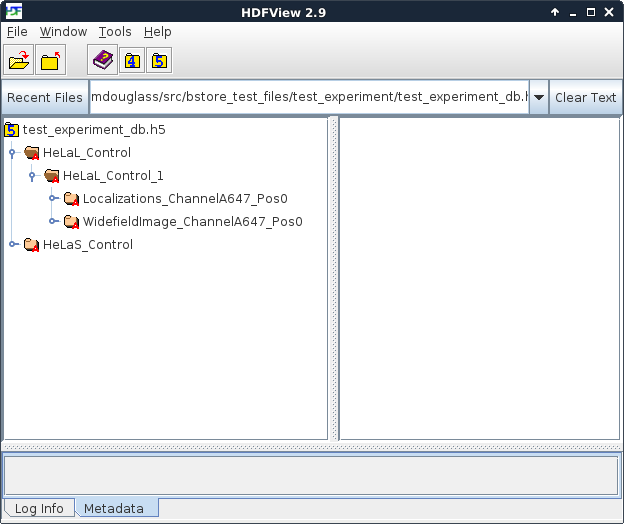
This group contains three different datasets: localizations (Localizations_ChannelA647_Pos0), a widefield image (WidefieldImage_A647_Pos0), and metadata describing how the localizations were obtained. (The metadata is not directly visible in this image because it’s stored as attributes of the Localizations_ChannelA647_Pos0 group.) Each dataset has two optional identifiers: a channelID of A647 and a posID of 0. The dataset keys–if they are specified–follow the format datasetType_channelID_posID_sliceID_dateID_replicateID. Because no sliceID, dateID, or replicateID is specified, they are absent from the name of the group.
Date ID’s are specified as strings in the format ‘YYYY-MM-DD’.
Position ID’s support single integer ID’s as one-tuples (0,) and two integer ID’s as two-tuples (1,4).
Example of a full HDFDatastore key¶
A HDFDatastore key using all the ID’s possible looks like:
HeLa_Control/HeLa_Control_76/Localizations_ChannelA750_Pos1_Slice5_Date20161211_Replicate5
The dataset IDs matching this key are prefix: HeLaControl, acqID: 76, datasetType: Localizations, channelID: A750, posID: 1, sliceID: 5, dateID: 20161211, replicateID: 5.
If posID was specified with two integers, such as (1,4), it the corresponding part of the key would look like Pos_001_004.
The dateID only has hyphens between the year, month, and day in Python; they are removed when writing to the HDF datastore.
As seen in the next figure, the actual localization data is stored as a table inside the Localizations_ChannelA647_Pos0 group. Metadata is attached as HDF attributes of the group; their values are in JSON format. Attributes have the same key as the dataset they belong to; if this dataset does not exist in the HDF file, neither will the metadata attributes. All attributes start with the string defined in the variable __HDF_Metadata_Prefix__ in config.py.
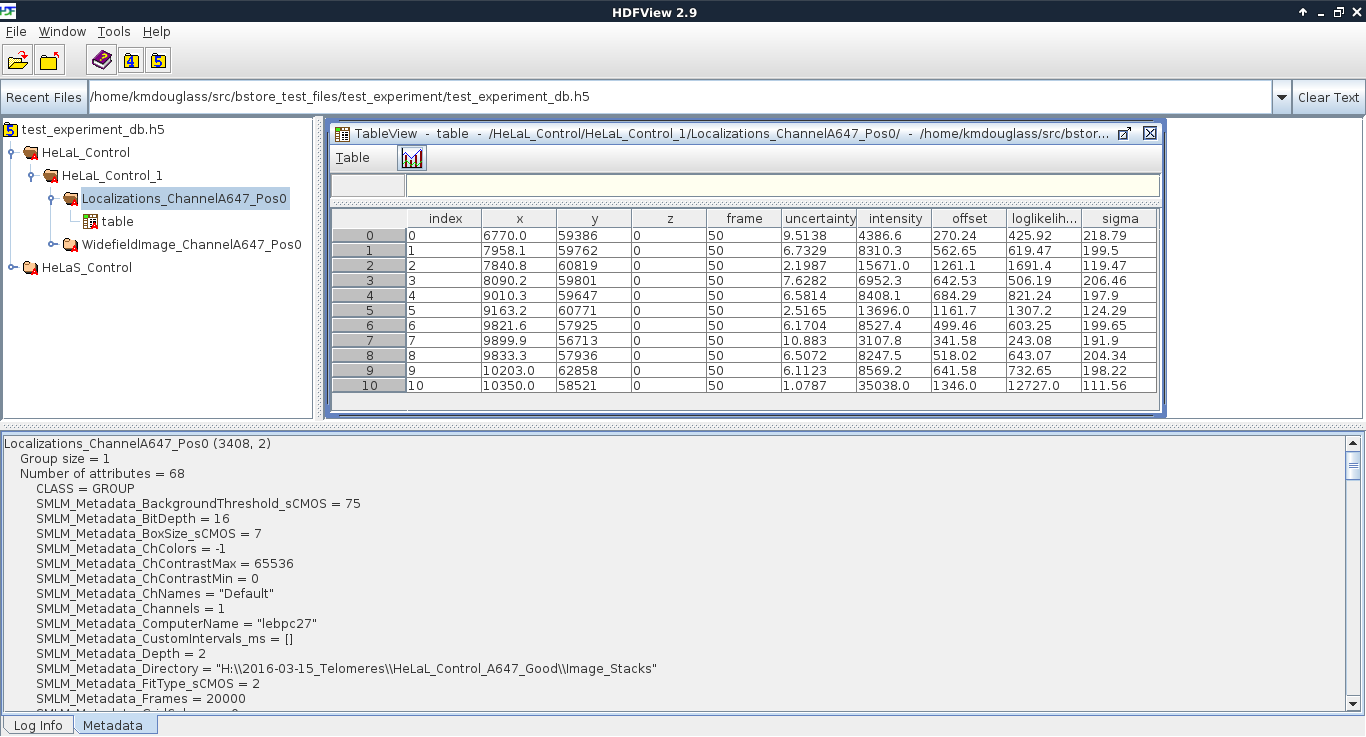
This mode of organization was chosen for a few reasons:
- The data is organized in a way that is easily read by both humans and machines. This means we can understand the organization of the data without any knowledge of how the datastore was created.
- B-Store dataset IDs can be inferred from the HDF key that points to the data. Machines can parse the HDF key to extract the dataset IDs, which is done, for example, when the function HDFDatastore.query() is executed.
- We take advantage of features provided by the HDF format, such as attributes and groups.